
Interphase: The Preparation for Cell Division
Interphase is a crucial phase of the cell cycle, representing the period during which a cell prepares for division. It is a time of growth, DNA replication, and preparation for mitosis, making it essential for the accurate duplication of genetic material and overall cell health. Interphase consists of three distinct phases: G1 (Gap 1), S (Synthesis), and G2 (Gap 2).
Overview of Interphase
Interphase is the longest part of the cell cycle, allowing the cell to grow, perform its normal functions, and prepare for division. It ensures that the cell is ready for the subsequent mitotic phase (M phase) and cytokinesis.
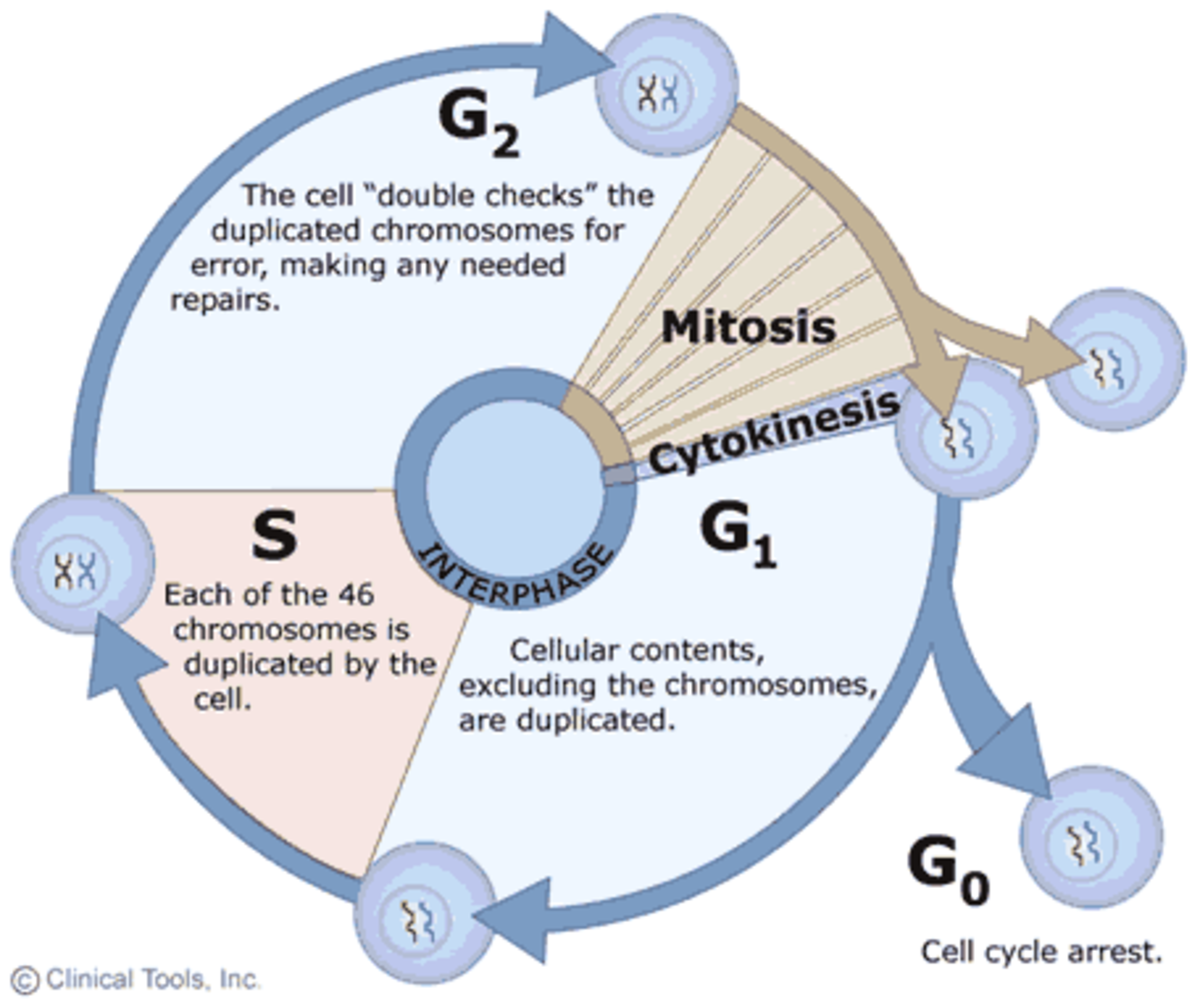
Phases of Interphase
G1 Phase (Gap 1)
The G1 phase is the first stage of interphase and focuses on cell growth and normal metabolic activities. During this phase, the cell:
- Grows in size: The cell increases its volume, synthesizes new proteins, and produces organelles. This growth ensures that each daughter cell will be of adequate size after division.
- Synthesizes mRNA and proteins: Essential proteins and mRNA necessary for DNA replication and cell cycle progression are synthesized.
- Monitors environment: The cell assesses whether conditions are favorable for division, such as the availability of nutrients, growth factors, and appropriate cell density.
G1 Checkpoint
The G1 checkpoint, also known as the restriction point, is a critical control mechanism that ensures the cell is ready to proceed to the S phase. It checks for:
- Cell size: The cell must be sufficiently large to support division.
- Nutrient availability: Adequate nutrients must be available to support cell growth and DNA synthesis.
- DNA integrity: Any DNA damage must be repaired before replication begins.
If conditions are not favorable, the cell may enter a quiescent state called G0, where it remains metabolically active but does not divide.
S Phase (Synthesis)
The S phase is dedicated to the replication of DNA. During this phase:
- DNA replication: Each chromosome is duplicated, resulting in two sister chromatids held together at the centromere. The entire genome is replicated to ensure that each daughter cell receives a complete set of genetic information.
- Histone production: Histones and other DNA-binding proteins are synthesized to package the newly replicated DNA into chromatin.
Replication occurs at multiple origins of replication along the DNA, ensuring efficient and timely duplication of the entire genome.
G2 Phase (Gap 2)
The G2 phase follows DNA synthesis and precedes mitosis. During this phase:
- Continued growth: The cell continues to grow and produce proteins necessary for mitosis.
- Organelle replication: Organelles such as mitochondria and the Golgi apparatus replicate to ensure that each daughter cell will have the necessary cellular machinery.
- Preparation for mitosis: Microtubules and other components required for chromosome segregation are synthesized.
G2 Checkpoint
The G2 checkpoint ensures that the cell is fully prepared for mitosis. It checks for:
- Complete DNA replication: Ensures that all DNA has been accurately replicated without any errors or damage.
- DNA damage repair: Any DNA damage detected must be repaired before the cell enters mitosis.
- Cell size: The cell must be large enough to support division.
If any issues are detected, the cell cycle is halted to allow for repair or completion of necessary processes.
| Feature | G1 Phase | S Phase | G2 Phase |
|---|---|---|---|
| Position in Cell Cycle | First gap phase, post-mitosis and pre-S phase | Synthesis phase, between G1 and G2 phases | Second gap phase, post-S phase and pre-mitosis |
| Primary Functions | Cell growth, protein synthesis, organelle production | DNA replication | Preparation for mitosis, DNA damage repair, protein synthesis |
| Key Regulatory Proteins | Cyclin D-CDK4/6, Cyclin E-CDK2 | Cyclin A-CDK2 | Cyclin A-CDK1, Cyclin B-CDK1 |
| Checkpoints | G1 checkpoint (restriction point) ensuring cell size, nutrients, growth signals, and DNA integrity | Intra-S phase checkpoint monitoring DNA damage and replication fork progression | G2/M checkpoint ensuring DNA replication completion and DNA damage repair |
| DNA Content | 2n (diploid), DNA not yet replicated | 2n to 4n transition, as DNA replication occurs | 4n (tetraploid), DNA replication complete |
| Duration | Variable, depending on cell type and conditions | Relatively constant, depending on organism and cell type | Relatively constant, shorter than G1 |
| Cell Cycle Commitment | Decision point for cell cycle entry (G0 or S phase) | Commitment to complete DNA replication | Commitment to mitosis if conditions are favorable |
| Major Cellular Activities | Metabolic activity, preparation for DNA replication | DNA synthesis and replication, histone production | Final preparations for mitosis, centrosome duplication |
| DNA Replication | No DNA replication; cell prepares for S phase | Active DNA replication, entire genome duplicated | DNA already replicated during S phase |
| Cell Size | Cell grows in size to prepare for DNA synthesis | Slight increase due to DNA synthesis and chromatin assembly | Cell continues to grow, reaching maximum size before mitosis |
Molecular Control of Interphase
Interphase progression is tightly regulated by cyclins and cyclin-dependent kinases (CDKs). Different cyclins and CDKs are active at various points of interphase:
- G1 cyclins: Bind to CDKs in the G1 phase to promote progression to the S phase.
- S-phase cyclins: Activate CDKs that initiate DNA replication.
- G2 cyclins: Bind to CDKs to prepare the cell for mitosis.
These cyclin-CDK complexes phosphorylate target proteins, driving the cell cycle forward and ensuring proper coordination of cell growth, DNA replication, and preparation for division.
Key Processes During Interphase
Interphase is not merely a period of dormancy but an active phase filled with critical biochemical and cellular activities. Here are some key processes that occur during each phase of interphase:
DNA Replication and Repair
During the S phase, DNA replication must be precise to ensure that each daughter cell inherits an exact copy of the genome. The replication process involves:
- Initiation: Replication begins at specific sites known as origins of replication. The replication machinery, including DNA helicases, unwinds the double helix to allow access to the DNA strands.
- Elongation: DNA polymerases synthesize new DNA strands by adding nucleotides complementary to the template strand. This process is semi-conservative, meaning each new double helix contains one old and one new strand.
- Termination: Replication continues until the entire genome is copied. Enzymes such as ligases seal any nicks in the sugar-phosphate backbone to complete the process.

During G2, the cell actively scans for and repairs any replication errors or DNA damage to prevent mutations and maintain genomic stability. Proteins involved in the DNA damage response, such as p53, play a pivotal role in halting the cell cycle and initiating repair mechanisms.
Protein Synthesis and Organelle Duplication
Interphase is marked by robust protein synthesis and organelle biogenesis to prepare the cell for division:
- Ribosome Production: Ribosomes are synthesized in the nucleolus during G1 and G2 phase to meet the increased demand for protein synthesis.
- Organelle Biogenesis: Organelles such as mitochondria and the Golgi apparatus duplicate to ensure each daughter cell inherits a full complement of cellular machinery. Mitochondria replicate through a process of fission, while the Golgi apparatus undergoes fragmentation and reassembly.
Metabolic Activities
The metabolic activities of the cell are heightened during interphase to provide the energy and substrates required for growth and replication:
- Glycolysis and Oxidative Phosphorylation: Increased metabolic flux through glycolysis and oxidative phosphorylation generates ATP, the energy currency of the cell.
- Nucleotide Synthesis: Sufficient nucleotides are synthesized to support DNA replication and RNA transcription.
- Lipid and Membrane Synthesis: Phospholipids and other components of cellular membranes are produced to support cell growth and the formation of new membranes during cytokinesis.
Cellular Checkpoints and Tumor Suppressors
Cell cycle checkpoints are critical control mechanisms that ensure the integrity and accuracy of cell division. Tumor suppressor proteins, such as p53 and retinoblastoma protein (Rb), are essential for these checkpoints:
- p53: Known as the "guardian of the genome," p53 responds to DNA damage by either arresting the cell cycle to allow for repair or inducing apoptosis if the damage is irreparable.
- Rb: The Rb protein controls the transition from G1 to S phase by regulating the activity of E2F transcription factors, which are necessary for the expression of genes involved in DNA replication.
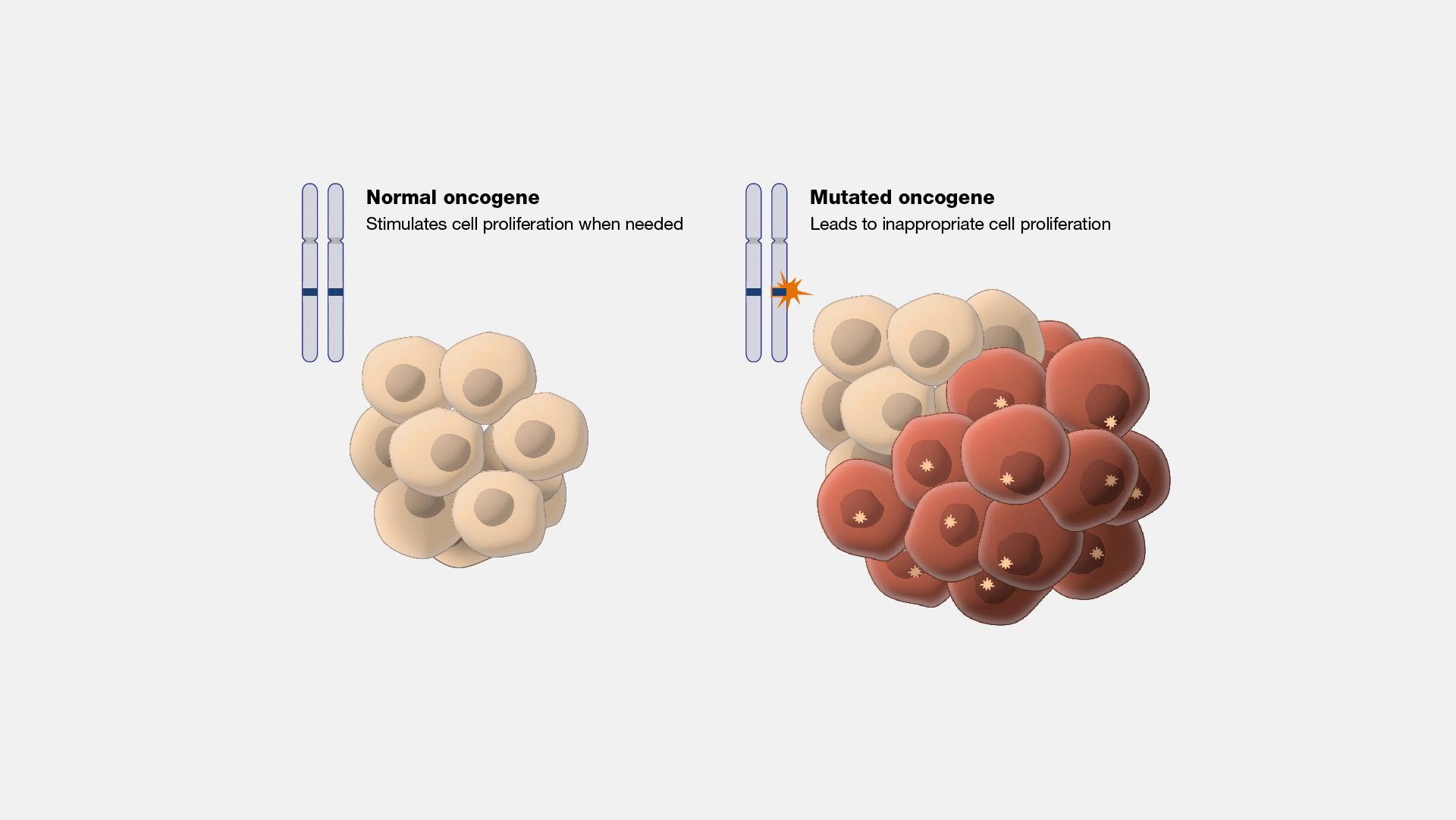
Implications of Interphase Dysregulation
Dysregulation of interphase processes can lead to numerous diseases, most notably cancer. Uncontrolled cell division, often due to mutations in genes that regulate the cell cycle (oncogenes and tumor suppressor genes), results in tumor formation and progression. Research into the molecular mechanisms governing interphase has led to targeted therapies aimed at correcting or exploiting these dysregulated pathways in cancer cells.
Advanced Research and Future Directions
Understanding interphase at a molecular level continues to be a rich field of research with implications for developmental biology, regenerative medicine, and cancer therapy. Current and future research focuses include:
- Epigenetic Regulation: Investigating how epigenetic modifications, such as DNA methylation and histone acetylation, regulate gene expression during interphase.
- Stem Cell Biology: Exploring how interphase regulation differs in stem cells, which must balance self-renewal with differentiation.
- Cancer Therapeutics: Developing drugs that specifically target cell cycle proteins to halt the proliferation of cancer cells without affecting normal cells.
Interphase FAQ
Epigenetic modifications, such as DNA methylation and histone acetylation, play crucial roles in regulating gene expression during interphase. These modifications can alter the accessibility of DNA to transcription factors and RNA polymerases, impacting the transcriptional activity of specific genes. Understanding the interplay between epigenetic regulators and gene expression dynamics during interphase is essential for unraveling complex cellular processes like development and disease.
Interphase regulation differs significantly between stem cells, which have the unique ability to self-renew and differentiate, and differentiated cells with more specialized functions. Stem cells often exhibit distinct patterns of gene expression and epigenetic modifications during interphase, allowing them to maintain pluripotency or multipotency. Understanding these differences can provide insights into stem cell biology, tissue regeneration, and potential applications in regenerative medicine.
Interphase is intimately linked to cellular senescence, a state of irreversible growth arrest associated with aging and age-related diseases. During interphase, various molecular pathways, including the p53-p21 and p16-Rb pathways, regulate cellular senescence by responding to stress signals, DNA damage, and telomere dysfunction. Senescent cells exhibit distinct morphological and molecular changes during interphase, such as enlarged cell size, altered chromatin organization, and increased expression of senescence-associated markers. Understanding the mechanisms underlying interphase-associated senescence is critical for elucidating the aging process and developing interventions to promote healthy aging.
During interphase, organelles grow and replicate to ensure equal distribution between daughter cells:
- Mitochondria: Increase in number through fission, regulated by proteins like Drp1 and Fis1.
- Endoplasmic Reticulum (ER) and Golgi Apparatus: Expand in size to meet the increased demands of the cell. The ER uses membrane-bound ribosomes for protein synthesis, and the Golgi modifies and sorts these proteins.
- Centrosomes: Duplicate once per cell cycle during the S phase, ensuring two centrosomes are available for mitotic spindle formation in mitosis.
Epigenetic modifications play a critical role in regulating gene expression during interphase:
-
DNA Methylation: Addition of methyl groups to DNA (usually at CpG islands) can silence gene expression. DNA methyltransferases (DNMTs) mediate this process.
-
Histone Modifications: Acetylation (by HATs) typically activates transcription, while methylation (by histone methyltransferases) can either activate or repress depending on the context. These modifications alter chromatin structure, making it more or less accessible to transcription machinery.
-
Non-Coding RNAs: Long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) regulate gene expression post-transcriptionally and through chromatin remodeling complexes.
Conclusion
Interphase is a critical phase of the cell cycle, encompassing the periods of growth, DNA replication, and preparation for mitosis. The G1, S, and G2 phases each play distinct roles in ensuring that the cell is ready for division, with checkpoints and regulatory mechanisms in place to maintain genomic integrity and cell health. Understanding interphase provides essential insights into cellular function, development, and the basis for many diseases, including cancer.
You can also read this article on the same topic https://www.toppr.com/guides/biology/cell-the-unit-of-life/what-is-interphase-definition-and-stages/
Also you can check out this article on Organization of chromatin in the interphase mammalian cell which explains how chromosomes behave throughout interphase in mitosis





